Background
Within the past century, biotechnology and molecular biology have seen remarkable progress as a result of explorations into anti-phage bacterial defense mechanisms, such as restriction enzymes and CRISPR-Cas systems. The discovery of restriction enzymes nearly sixty years ago opened the door to precise cutting and manipulation of DNA sequences, thus marking the birth of modern genetic engineering. The accessibility of genome editing ramped up decades later with the development of CRISPR-Cas systems as gene editing tools, revealing unparalleled precision and versatility.
Anti-phage bacterial defense systems have led to some of the most intriguing scientific breakthroughs in the world of biotechnology, from synthetic biology to gene therapies. Despite this, hundreds of additional phage defense systems remain unexplored.
Defense-Associated Reverse Transcriptases
Reverse transcriptases (RTs) are enzymes that catalyze the polymerization of DNA from an RNA template. Defense-associated reverse transcriptases (DRTs) are novel reverse transcriptases involved in bacterial anti-phage defense, however the mechanisms by which DRTs confer anti-phage defense are largely unknown. Characterization of these pathways may elucidate potential diverse biotechnological applications.
DRT2
Recently the function of the defense-associated reverse transcriptase 2 (DRT2), has been discovered. The genes encoding for DRT2 consist of only two components: an RT domain and a noncoding RNA (ncRNA). These minimal systems are sufficient to mount an abortive infection (Abi) response in order to defend against foreign invaders.
Within the defense pathway undertaken by DRT2 a portion of the ncRNA is converted into ssDNA by the RT through rolling-circle reverse transcription (RCRT), resulting in the formation of long cDNA products with concatenated repeats. This cDNA product contains an open-reading-frame (ORF) that lacks any in-frame stop codons and is thus referred to as neo (nearly endless ORF). When under phage infection, the single-stranded cDNA undergoes second-strand synthesis to become dsDNA, which is then transcribed into concatenated mRNA. Following this, the mRNA is translated into a Neo protein which leads to programmed cell dormancy, the mechanism by which the cell defends against bacteriophages.
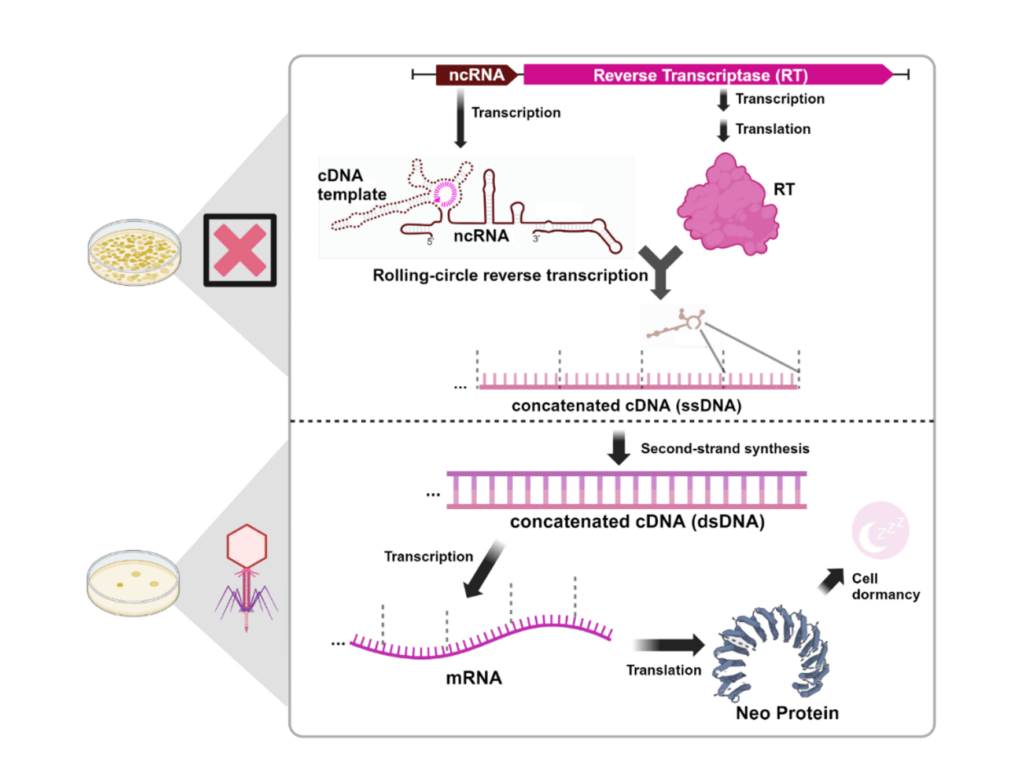
This defense mechanism highlights novel coding potential in the genome by generating genes from RNA templates, challenging the traditional understanding of how genetic information is stored.
Discussion
While it has been demonstrated that the functional outcome of Neo expression is cell growth arrest, the biological pathway connecting Neo expression to growth arrest remains unclear. Through continuing to pursue the investigation of the Neo protein and its role in the DRT2 defense mechanism, science could gain further insights into newfound complexities in the central dogma of molecular biology may aid in opening the door for future potential medical applications
Resources
Loenen W. et al. (2014) Highlights of the DNA cutters: a short history of the restriction enzymes, Nucleic Acids Res. 42(1):3-19
Jinek M. et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity, Science. 337(6096):816-21.
Georjon H, Bernheim A. (2023) The highly diverse antiphage defence systems of bacteria, Nat Rev Microbiol. 21(10):686-700.
Gonzalez-Delgado A. et al. (2021) Prokaryotic reverse transcriptases: from retroelements to specialized defense systems, FEMS Microbiol Rev. 45(6)
Tang S. et al. (2024) De novo gene synthesis by an antiviral reverse transcriptase, bioRxiv 2024.05.08.593200; doi: https://doi.org/10.1101/2024.05.08.593200
Disclaimers
Our articles are based on works by board-certified/accredited doctors and medical researchers. We do not claim to be a professional peer-reviewed publication. This article was not written by medical professionals.