Disclaimer: This article was not written by medical professionals. If you suspect any signs of skin cancer, please contact your dermatologist.
Introduction
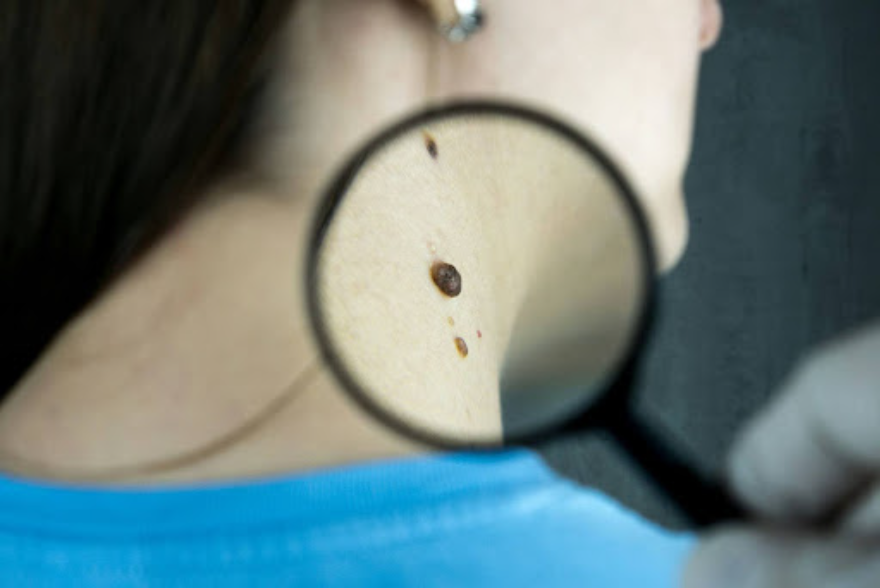
Skin cancer is the most common type of cancer worldwide, accounting for one-third of all cancer diagnoses. Fortunately, skin cancer is generally easy to diagnose and, when caught early, is very treatable, with a low mortality rate.
Skin cancer can be broken down into two main categories: melanoma skin cancer and non-melanoma skin cancer. Non-melanoma skin cancer can be further broken down into basal cell carcinoma (BCC) and squamous cell carcinoma (SCC). According to Mayo Clinic, risk factors include excessive sun exposure, old age, radiation, having a fair complexion, immunosuppression, and a personal or family history of skin cancer.
Basal cell carcinoma is the most prevalent form of skin cancer and also the least dangerous due to its low probability of metastasizing. Squamous cell carcinoma occurs more often than malignant melanoma, however, it tends to be less aggressive and less invasive. All three forms of skin cancer can be diagnosed by a pathologist viewing a skin biopsy taken by a dermatologist.
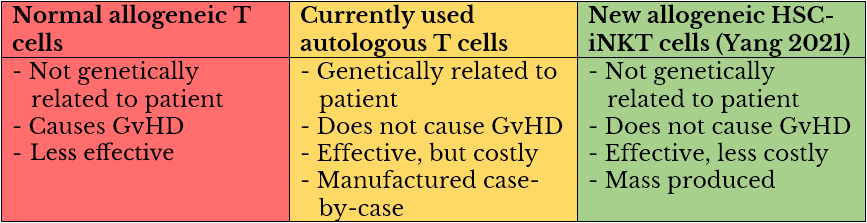
Basal Cell Carcinoma
Basal cell carcinoma is the most common form of skin cancer, and is often the easiest to identify. BCC can look like a smooth waxy bump on the skin. Pain, tenderness, itching, and bleeding are all signs that indicate a bump needs to be examined by a dermatologist. Some patients also report persistent pimples that don’t go away, which can be a warning sign of basal cell carcinoma.
Squamous Cell Carcinoma
Most squamous cell carcinomas begin as rough, raised, dry spots called actinic keratoses. Medical professionals can treat these lesions with liquid nitrogen, topical fluorouracil cream, or red light therapy. If left untreated, actinic keratoses can develop into SCC. Skincancer.org states that SCC can look like “a persistent, scaly red patch with irregular borders that sometimes crusts or bleeds.”
Malignant Melanoma
Melanoma presents as one or multiple dark spots on the skin. This type of cancer expands rapidly and can metastasize to the lymph nodes and other organs. To differentiate melanomas from other skin spots, The American Academy of Dermatology developed the ABCDE memory mnemonic: asymmetry, border, color, diameter, and evolving. Following their guidelines, if a dark spot is asymmetrical, has jagged borders, various color shades, a diameter larger than a pencil eraser, or is changing in any way, it may indicate a malignant melanoma, and should be evaluated by a dermatologist.
Treatment
There are many approaches to skin cancer prevention, but the most feasible is adopting measures like wearing sunscreen with at least 30 SPF daily (and reapplying every two hours) or simply avoiding significant sun exposure.
Skin cancer treatment begins with a visit to a medical professional who can perform a biopsy on the concerning area. Once skin cancer is diagnosed after a biopsy, treatment typically involves surgery. In some cases, mild forms of squamous cell carcinoma called SCC in situ (superficial squamous cell carcinoma that has not spread) can be treated with liquid nitrogen therapy. In other cases, skin cancers will have to be excised (removed surgically). If the cancer is located in a place with limited skin to work with, such as the face or nose, the patient may opt for a Mohs surgery. Mohs surgery is a tissue-sparing surgery technique that takes place in stages. The physician will operate in stages taking as little tissue as possible and looking at it under a microscope. If there is more cancer under the microscope, they will continue with another stage, and this process is repeated until all margins are clear and the patient is cancer-free with as little skin missing as possible. According to Dr. Tolkachjov, medical professionals do not begin reconstruction until after all the margins are confirmed to be histologically tumor-free.
References
What to look for: Abcdes of melanoma. American Academy of Dermatology. (n.d.). https://www.aad.org/public/diseases/skin-cancer/find/at-risk/abcdes
Mayo Foundation for Medical Education and Research. (n.d.). Basal cell carcinoma. Mayo Clinic. https://www.mayoclinic.org/diseases-conditions/basal-cell-carcinoma/symptoms-causes/syc-20354187
professional, C. C. medical. (n.d.). Melanoma: Symptoms, staging & treatment. Cleveland Clinic. https://my.clevelandclinic.org/health/diseases/14391-melanoma
Squamous cell carcinoma warning signs and images. The Skin Cancer Foundation. (2023, January 9). https://www.skincancer.org/skin-cancer-information/squamous-cell-carcinoma/scc-warning-signs-and-images/
Tolkachjov, S. N., Brodland, D. G., Coldiron, B. M., Fazio, M. J., Hruza, G. J., Roenigk, R. K., Rogers, H. W., Zitelli, J. A., Winchester, D. S., & Harmon, C. B. (2017). Understanding mohs micrographic surgery. Mayo Clinic Proceedings, 92(8), 1261–1271. https://doi.org/10.1016/j.mayocp.2017.04.009 WebMD. (n.d.). Skin cancer: Melanoma, basal cell, and squamous cell carcinoma. WebMD. https://www.webmd.com/melanoma-skin-cancer/skin-cancer