During the pandemic, laboratories across the world worked hard to improve current diagnostic testing methods. The main method, quantitative polymerase chain reaction (qPCR) turns a small quantity of DNA into a larger amount and uses fluorescent dyes to indicate the presence or absence of viral genetic material. However, this method falls short in the following ways:
- It requires expensive equipment and reagents, along with trained personnel.
- It requires temperature cycles, so it cannot be performed at a single temperature.
- It takes time. Depending on the initial amount of target present, a qPCR test can take as long as 90 minutes.
At the University of Florida, PhD student Long T. Nguyen, working under Dr. Piyush Jain, has developed a rapid, single temperature COVID-19 diagnostic test that provides results in under 30 minutes. Amazingly, the test distinguishes between five COVID variants, achieves amplification, and RNA to DNA conversion all in one “pot.” Finally, the results can be read on a mobile phone.
The system they used is based on a detection system found in bacteria. Bacteria contain natural immune systems called CRISPR Cas, which function to create both a memory of past viral infections, along with a defense system once these viruses come back. Cas is a protein which is sometimes described as “a pair of molecular scissors,” capable of cutting DNA or RNA fragments, while CRISPR contains complementary sequences to attacking viruses and acts as a “molecular GPS,” helping Cas find a certain target. For this reason, it is also called a Guide RNA.

Some Cas proteins locate their target and only make cuts around the target DNA/RNA; this is called cis cleavage. Others go on a “cutting frenzy.” After finding the target and cutting, the Cas protein starts cutting up other DNA or RNA fragments surrounding it, termed trans cleavage. Cas9 proteins, famous for genetic engineering, employ cis cleavage and only cut DNA. Cas12 and Cas13 proteins utilize trans cleavage, cutting DNA and RNA respectively. All bacteria have adapted their own systems, with slight variations, allowing scientists to harness each’s individual powers.
Cas12 and Cas13 proteins are at the forefront of diagnostic research. Their cutting frenzy may not be great for gene editing, but recent innovation has found that FQ reporters, or fluorescent quenchers, can be used to detect a signal with light. These reporters are dampened by a piece of RNA or DNA located between the fluorescent and quencher. Once cut, they glow and show a light on a fluorescent reader. If a Cas12 or Cas13 protein detects its target, it will cut the target then rapidly start cutting the FQ reporters nearby.
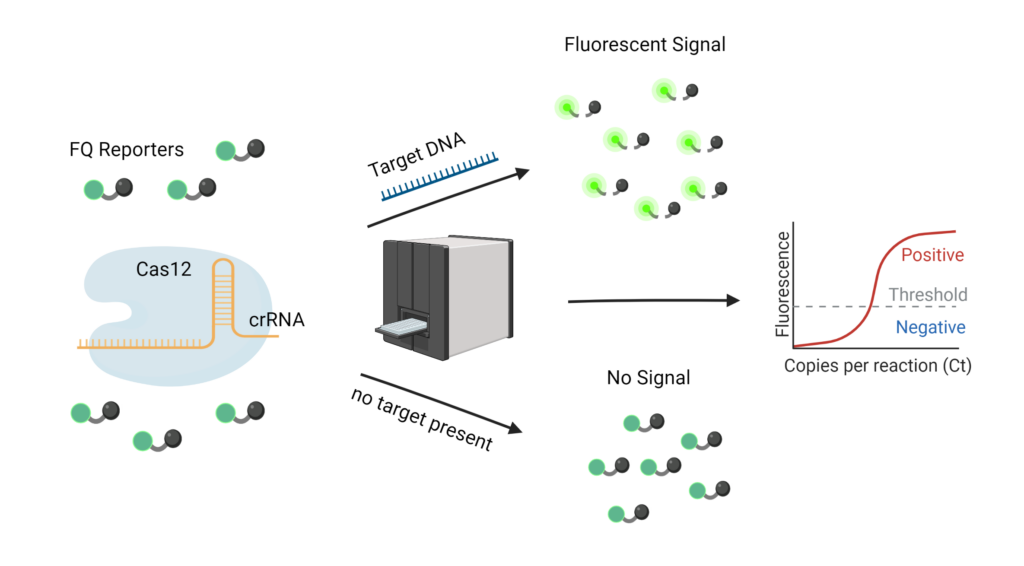
CRISPR RNA, shortened as crRNA, can be “programmed” to target any part of a target sequence. Specifically for the virus that causes COVID-19, there is a highly conserved region called the N gene. Since this same sequence is found across all variants, it can indicate the presence of the virus, but does not distinguish between mutated strains. The Jain lab identified mutated regions on each of the five variants: Alpha, Beta, Gamma, Delta, and Omicron, and created crRNAs which were complementary to each of these mutated regions.
They then had to choose the optimal Cas protein, which could withstand higher temperatures. This was necessary because amplification occurs at high temperatures, ranging from 55-70°C. BrCas12b comes from a thermophilic bacterium found in hot springs and was the optimal choice.
They combined this Cas protein, along with a crRNA and finally, a master mix of RT-LAMP (Reverse Transcription Loop Mediated Isothermal Amplification). This is a very complex sounding term, but it can be broken down fairly easily. Reverse transcription is the process of converting RNA into DNA. Isothermal means it works at a single temperature and amplification implies the amount of DNA increases greatly. This amplification also provides a checkpoint. Researchers were able to first see if the patient sample was amplified; if so, COVID must be present. Then, using the CRISPR Cas system, they can determine exactly what variant is present.
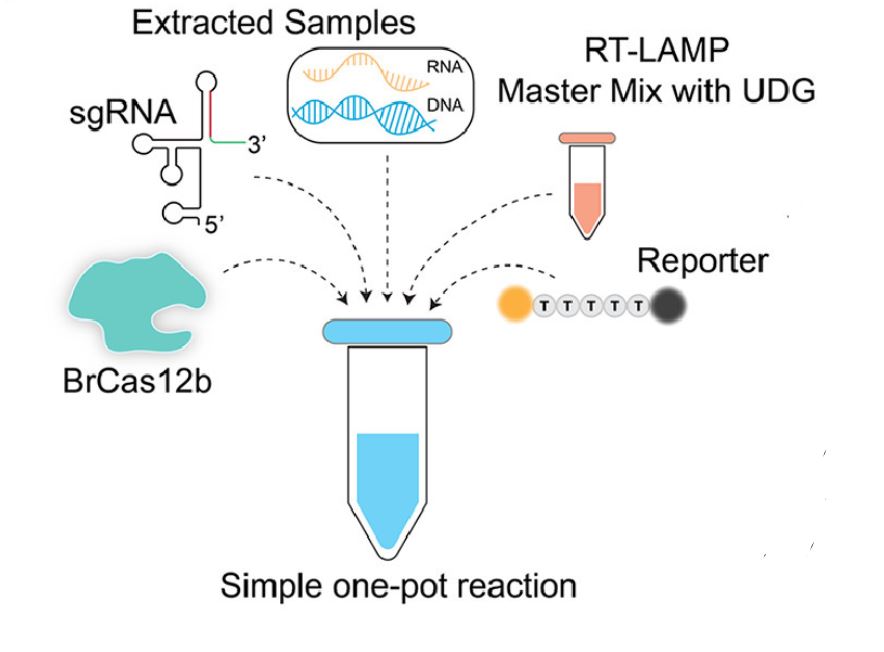
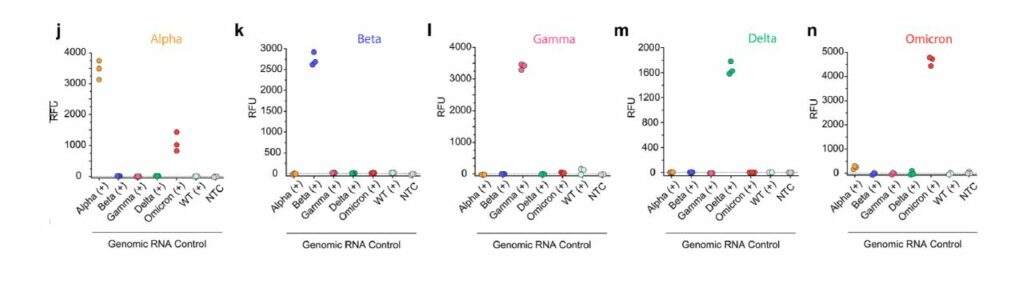
Detection is completed in under 30 minutes and patient samples with a higher viral load (i.e., they had more SARS-CoV-2 virions present in their sample), exhibited 100% accuracy with about 95% sensitivity in distinguishing variants. The figure below, from Long et. al shows the incredible accuracy which comes from this detection. The colored titles, “Alpha, Beta, etc.” are the variant present in the patient sample, while the x-axis shows the crRNA used. For instance, it was expected that if a patient contained the Beta strain, only the Beta crRNA would show high signal.
Most studies combining RT-LAMP with a CRISPR reaction have extremely low sensitivity and difficulty distinguishing a positive sample. The use of specifically BrCas12b, a less studied Cas protein, allowed the Jain lab to circumvent many of the problems others have had combining the two. The applicability of this research extends far beyond COVID-19 detection. Any RNA or DNA detection could be done utilizing this research, simply by changing the sequence located on the crRNA.
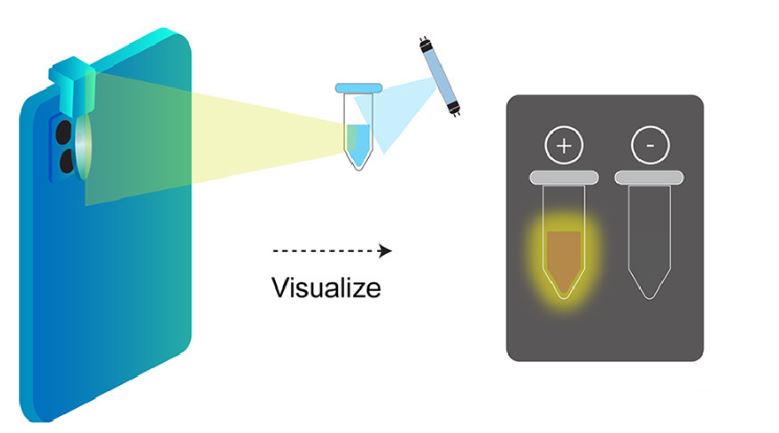
Moreover, the Jain lab aims to create portable and cheap methods for testing. They proposed an inexpensive lens which can be attached to any mobile phone camera. In a dark setting, the lens, which costs less than $5 can shine light of a specific wavelength on a sample with the added CRISPR Cas reagent and glow in the presence of COVID-19.
References
- Kubina, R., & Dziedzic, A. (2020, June 26). Molecular and serological tests for COVID-19. A comparative review of SARS-Cov-2 coronavirus laboratory and point-of-care diagnostics. MDPI. https://doi.org/10.3390/diagnostics10060434
- Nguyen, L. T., et al. (2022, March 1). A Thermostable Cas12b from Brevibacillus Leverages One-pot Detection of SARS-CoV-2 Variants of Concern. eBioMedicine, The Lancet Discovery Science. https://doi.org/10.1016/j.ebiom.2022.103926